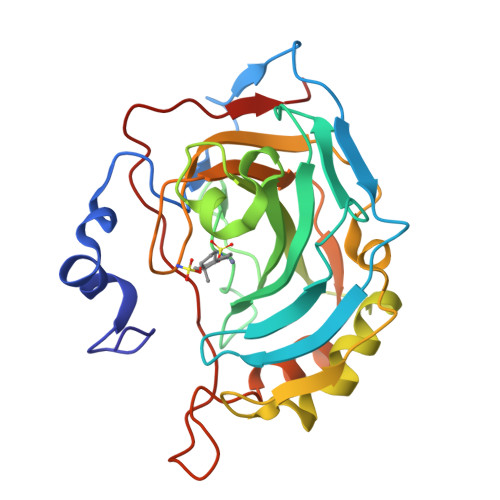
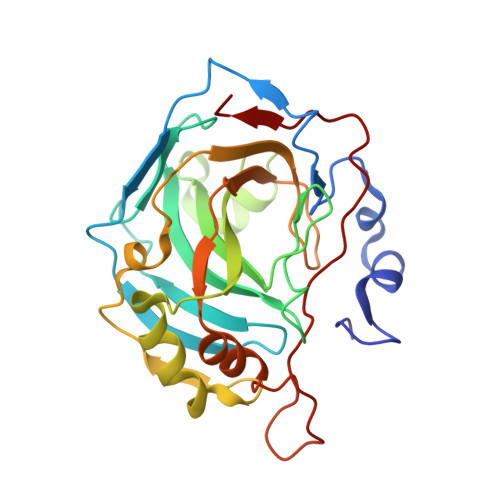
3,17 beta-Bis-sulfamoyloxy-2-methoxyestra-1,3,5(10)-triene and Nonsteroidal Sulfamate Derivatives Inhibit Carbonic Anhydrase IX: Structure-Activity Optimization for Isoform Selectivity.
Andring, J.T., Dohle, W., Tu, C., Potter, B.V.L., McKenna, R.(2019) J Med Chem 62: 2202-2212
- PubMed: 30721041
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01990
- Primary Citation of Related Structures:
6E8P, 6E8X, 6E91, 6E92 - PubMed Abstract:
3,17β-Bis-sulfamoyloxy-2-methoxyestra-1,3,5(10)-triene (STX140), a bis-sulfamate derivative of the endogenous steroid 2-methoxyestradiol, has shown promising anticancer potency both in vitro and in vivo, with excellent bioavailability. Its activity against taxane-resistant xenografts makes it a potential drug candidate against triple-negative breast cancer (TNBC). These properties are linked to the ability of STX140 to act in a multitargeting fashion in vivo as a microtubule disruptor, leading to cell cycle arrest and with both proapoptotic and anti-angiogenic activities. Carbonic anhydrase IX (CA IX) is a well-established biomarker for aggressive cancers, including TNBC. This study reports, for the first time, the inhibitory activities of a series of steroidal and nonsteroidal sulfamate derivatives against CA IX in comparison to the ubiquitous CA II, with some compounds demonstrating 100-200-fold selectivity for CA IX over CA II. X-ray crystallographic studies of four of the most promising compounds reveal that isoform-specific residue interactions are responsible for the high specificity.
Organizational Affiliation:
College of Medicine, Department of Biochemistry and Molecular Biology , University of Florida , Gainesville , Florida 32610 , United States.